The Ghent University Digital PCR Center (DIGPCR)
DIGPCR combines the complementary expertise from various Ghent University researchers on digital PCR. We collaborate with internal and external partners on using digital PCR for various applications. As expert users of this technology, we frequently perform trainings on dPCR and give lectures at conferences or workshops. We are also co-authors on the dMIQE guidelines for transparent reporting of dPCR data.

A better way to start building.
Equipment
Our center has access to different dPCR platforms, read about our equipment, where to find it and who to contact.
Software
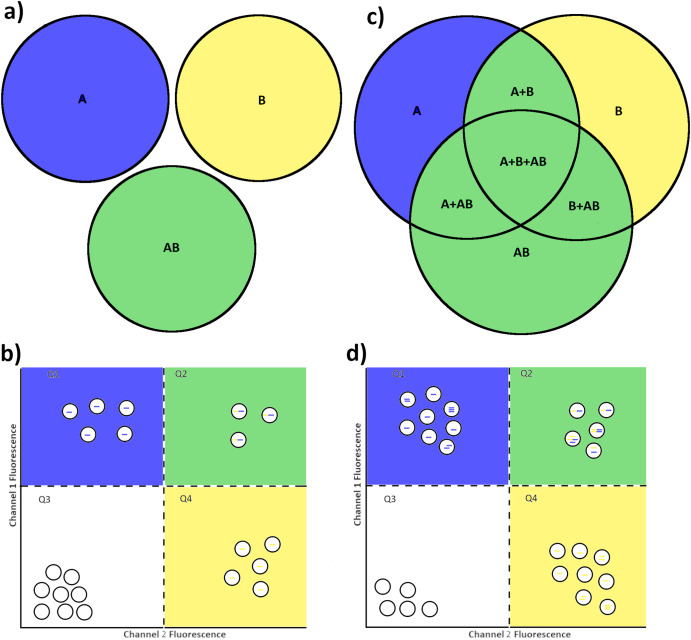
Use the software tools we have developed for dPCR analysis, including tresholding methods, assay validation methods and power calculation for dPCR experiments.
Events and Courses
Learn about our yearly dPCR course, our congress contributions, or view previuos contributions.
News and Events in the spotlight

Nio™+ early access experience
Learn our experience as early access users of the Nio™+ by Stilla technologies

SC2014
dPCR specialist course
Our yearly digital PCR specialist course will be organized from 10-12 Septmber 2024 at Campus Merelbeke, Belgium. Click here for more information